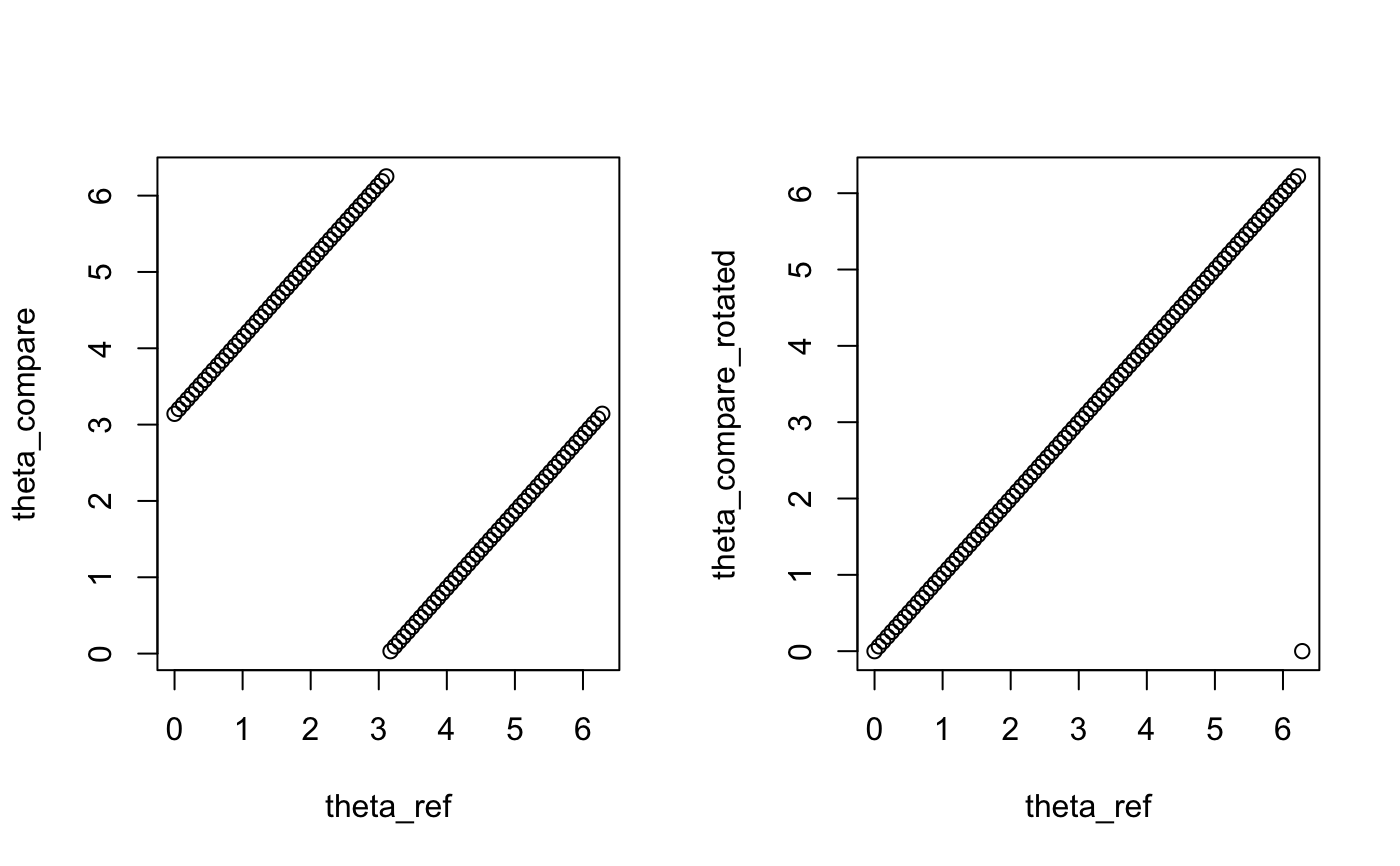
Rotate circular variable shift_var to minimize
the distance between ref_var and shift_var. Because
the origin of the cell cycle phases is arbitrary, we transform the
angles prior to computing the distance (rotation and
shifting). After this, one can apply circ_dist to compute
the distance between the output value and ref_var.
rotation(ref_var, shift_var)
Arguments
| ref_var | A vector of reference angles. |
|---|---|
| shift_var | A vector of angles to be compared to ref_var. |
Value
The transformed value of shift_var after rotation and
shifting.
Examples
# Create a vector of angles. theta_ref <- seq(0,2*pi, length.out=100) # Shift the origin of theta_ref to pi. theta_compare <- shift_origin(theta_ref, origin = pi) # Rotate theta_compare in a such a way that the distance between # theta_ref and theta_compare is minimized. theta_compare_rotated <- rotation(ref_var=theta_ref, shift_var=theta_compare) par(mfrow = c(1,2)) plot(x=theta_ref, y = theta_compare) plot(x=theta_ref, y = theta_compare_rotated)